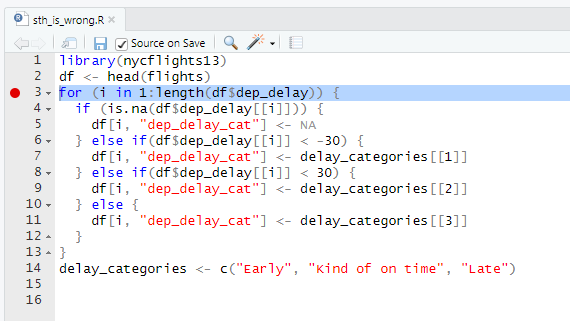
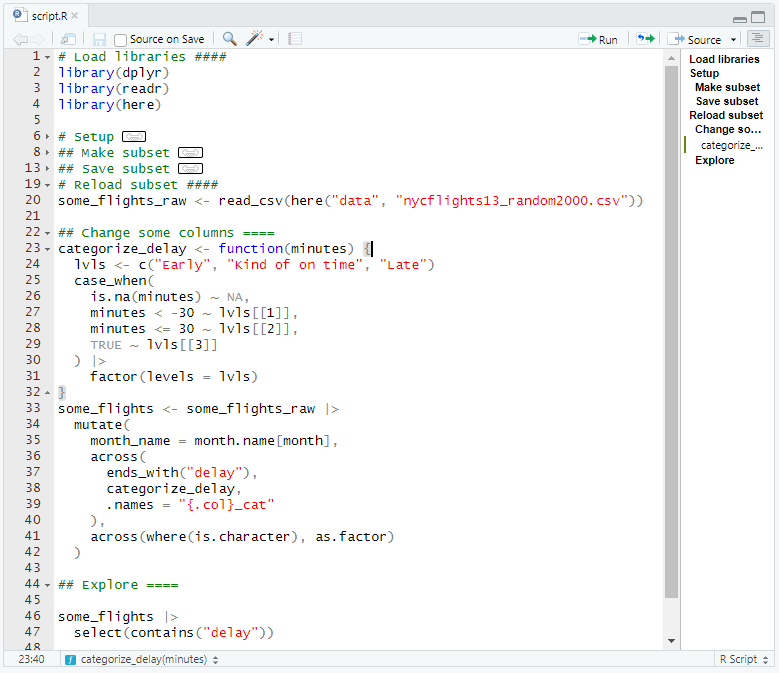
[1] "carrier,flight,month_name,time_hour,dep_delay,arr_delay,dep_delay_cat,arr_delay_cat"
[2] "UA,1168,April,2013-04-12T22:00:00Z,61,33,Late,Late"
[3] "VX,407,February,2013-02-28T14:00:00Z,-1,-50,Kind of on time,Early"
[4] "DL,1174,August,2013-08-22T15:00:00Z,0,-6,Kind of on time,Kind of on time"
[5] "UA,1722,July,2013-07-31T10:00:00Z,5,-12,Kind of on time,Kind of on time"
[6] "B6,1801,September,2013-09-11T20:00:00Z,-2,-10,Kind of on time,Kind of on time"
[7] "EV,4212,January,2013-01-06T20:00:00Z,4,4,Kind of on time,Kind of on time"
[8] "UA,671,August,2013-08-26T11:00:00Z,-7,-31,Kind of on time,Early"
[9] "EV,4567,December,2013-12-23T15:00:00Z,14,28,Kind of on time,Kind of on time"
[10] "WN,165,July,2013-07-18T17:00:00Z,12,-5,Kind of on time,Kind of on time"