Best practices for R on HPC
2024-12-03
Overview
- Motivation
- How to run R on HPC systems
- Open OnDemand
- Batch jobs
- Walltime & memory
- Efficiency
- Improving performance
- Profiling
- Code optimization
- Wrap up
Motivation
Why R on HPC?
- Computation takes lots of time
- Large dataset, not enough RAM
- Many different scenarios to compute
Or any combination of these
Is it hard?
Warning
There is a learning curve
Tip
It’s not rocket science
- Training is available
- Support is there to help
Running R on HPC systems
Open OnDemand
- Web-based platform for accessing HPC resources
- RStudio
- Jupyter Lab
- Terminal
- File browser
- Convenient for interactive development
- Very inefficient for computations
Access via https://ondemand.hpc.kuleuven.be/
Batch jobs
Example “computation” script:
hello_world.R
No suprises here
Testing the script
Terminal:
- 1
- Load the R version you want to use
- 2
- Run the script
Warning
Don’t run computationally intensive tasks on the login node!!!
Module system
What & why?
- Different versions of software packages
- Reproducibility
How?
- Search for modules:
module -r spider ^R/ - Load a module:
module load R/4.4.0-gfbf-2023a - List loaded modules:
module list - Unload all modules:
module purge
Job script
hello_world.slurm
- 1
- Initialize the environment
- 2
- Specify the account for credits
- 3
- Number of nodes
- 4
- Number of tasks
- 5
- Number of CPUs per task
- 6
- Maximum runtime of job
- 7
- Cluster to run on
- 8
- Unload all modules
- 9
- Load the R version
- 10
- Run the script
Slurm speak
Slurm is
- resource manager
- job scheduler
User(s) submit jobs to Slurm
- Job in queue
- When resources available, job starts
Efficient use of (very expensive) HPC resources
Submitting a job
- 1
-
Submit the job script
hello_world.slurm - 2
-
Job ID is
62136541(incremental)
Checking job status
Show all your jobs on cluster wice:
ST is the job state:
PD: pendingR: runningCG: completing
No jobs shown? All done!
Job output
Saved to file slurm-<jobid>.out:
slurm-62136541.out
SLURM_JOB_ID: 62136541
SLURM_JOB_USER: vsc30140
SLURM_JOB_ACCOUNT: lpt2_sysadmin
SLURM_JOB_NAME: hello_world.slurm
SLURM_CLUSTER_NAME: wice
SLURM_JOB_PARTITION: batch
SLURM_NNODES: 1
SLURM_NODELIST: p33c20n3
SLURM_JOB_CPUS_PER_NODE: 1
Date: Tue Aug 6 08:58:09 CEST 2024
Walltime: 00-00:02:00
========================================================================
Hello from p33c20n3 !- 1
- Info about job
- 2
- Output of the script
Runtime parameters
Scenario: run R script with different parameters
optparse package
hello_world_cla.R
library(optparse)
# Define the options
option_list <- list(
make_option(c("--name"), type="character", default="World",
help="Name to greet"),
make_option(c("--count"), type="integer", default=1,
help="Number of times to greet")
)
opt <- parse_args(OptionParser(option_list=option_list))
for (i in 1:opt$count) {
cat("Hello", opt$name, "!\n")
}- 1
-
Load the
optparsepackage - 2
- Define the options
- 3
-
Option for the name,
charactertype, default value “World” - 4
-
Option for the count,
integertype, default value 1 - 5
- Parse the arguments
- 6
-
Use the arguments in list
opt
Job script with parameters
File I/O
Easiest option: relative paths
$ Rscript ./weather_analysis.R \
--input data/London.csv \
--quantity temp_max \
--output tempmax_london.csv- 1
-
Input file in
datadirectory, subdirectory of script’s directory - 2
- Output file in script’s directory
Warning
Paths are relative to location you run script from!
Here: directory that contains weather_analysis.R
R script for weather analysis
weather_analysis.R
library(optparse)
option_list <- list(
make_option(c("-i", "--input"), type = "character",
help = "Path to the input CSV file"),
make_option(c("-q", "--quantity"), type = "character",
help = "Name of the quantity to analyze (e.g., temp, tempmax, tempmin)"),
make_option(c("-o", "--output"), type = "character",
help = "Path to the output CSV file")
)
opt <- parse_args(OptionParser(option_list = option_list))
data <- read.csv(opt$input)
if (!(opt$quantity %in% colnames(data))) {
stop(paste("The specified quantity", opt$quantity, "is not present in the data"))
}
data$month <- format(as.Date(data$datetime), "%m")
monthly_avg <- aggregate(data[[opt$quantity]], by = list(data$month), FUN = mean)
colnames(monthly_avg) <- c("Month", "Average")
write.csv(monthly_avg, file = opt$output, row.names = FALSE)Job script for weather analysis
weather_analysis.slurm
#!/usr/bin/env -S bash -l
#SBATCH --account=lpt2_sysadmin
#SBATCH --time=00:02:00
#SBATCH --cluster=wice
# first clean up your environment, then load R
module purge &> /dev/null
module load R/4.4.0-gfbf-2023a
# define output directory and create it if it does not exist
OUTDIR="output"
mkdir -p $OUTDIR
# define the quantity to analyse
QUANTITY="tempmax"
Rscript ./weather_analysis.R \
--input data/London.csv \
--quantity "$QUANTITY" \
--output $OUTDIR/$QUANTITY.csvBut wait, there is no optparse!
Many available through
R-bundle-CRANmoduleNot all packages are installed though
Install packages in
VSC_DATAdirectory- Pure R/no depencies on external libraries: easy
- Other packages: more difficult, use modules
Packages specific for R major version/cluster/architecture
Installation procedure
Start interactive job on cluster wice
Prepare environment
Define environment variable
Create package directory for cluster/architecture/R version
Only once per cluster/architecture/R version!
Actual installation
Install the package(s)
Tip
Use renv for managing R environments and dependencies
If you have trouble, contact the HPC helpdesk, specify
- packages to install
- R version
- cluster and architecture
Walltime & memory
How long will your job run?
- Specify walltime (
--time=HH:MM:SS) - But how much?
- Don’t underestimate!
- Don’t (massively) overestimate
- Experiment
- Start with “small” problem
- Increase problem size gradually
- Extrapolate to real problem
Running example
R script dgemm.R
- takes argument \(N\) and \(p\)
- generates matrix \(A \in \mathbb{R}^{N \times N}\), elements normally distributed
- computes \(A^p\)
- determines minimum and maximum diagonal element
Benchmark job script
memory_and_walltime.slurm
#!/usr/bin/env -S bash -l
#SBATCH --account=lpt2_sysadmin
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --time=01:00:00
#SBATCH --partition=batch_sapphirerapids
#SBATCH --cluster=wice
module load R/4.4.0-gfbf-2023a
module load GCCcore/12.3.0
Rscript dgemm.R --size $SIZE --power $POWER- 1
-
Note
SIZEandPOWERenvironment variables
Running benchmark
Submit with range of values for SIZE and POWER, e.g.,
$ sbatch --export=ALL,SIZE=5000,POWER=10 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=5000,POWER=20 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=5000,POWER=40 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=10000,POWER=10 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=10000,POWER=20 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=10000,POWER=40 memory_and_walltime.slurm
...Walltime benchmark results
| job ID | \(N\) | \(p\) | walltime |
|---|---|---|---|
| 62199204 | 5000 | 10 | 00:02:17 |
| 62199205 | 5000 | 20 | 00:04:36 |
| 62199206 | 5000 | 40 | 00:08:50 |
| 62199207 | 10000 | 10 | 00:15:10 |
| 62199208 | 10000 | 20 | 00:30:57 |
| 62199209 | 10000 | 40 | 01:00:15 |
| 62199210 | 20000 | 10 | 00:04:19 |
| 62199211 | 20000 | 20 | 00:04:24 |
| 62199212 | 20000 | 40 | 00:04:34 |
| 62199213 | 50000 | 10 | 00:00:55 |
| 62199214 | 50000 | 20 | 00:00:45 |
| 62199215 | 50000 | 40 | 00:00:44 |
- \(T(N, p) \propto p\)
- \(T(N, p) \propto N^3\)
- What happened for \(N = 10000\), \(p = 40\)?
- What happend for \(N = 20000\) and \(N = 50000\)?
Out of time
Inspect output for job 62199209
...
Walltime: 00-01:00:00
========================================================================
Lmod is automatically replacing "cluster/genius/login" with
"cluster/wice/batch_sapphirerapids".
slurmstepd: error: *** JOB 62199209 ON q16c03n1 CANCELLED
AT 2024-08-13T18:28:16 DUE TO TIME LIMIT ***- 1
- Oopsie: walltime exceeded requested walltime
Warning
No output, 1 hour of computime lost!
What about \(N\) = 20000, 50000
| job ID | \(N\) | \(p\) | walltime |
|---|---|---|---|
| 62199210 | 20000 | 10 | 00:04:19 |
| 62199211 | 20000 | 20 | 00:04:24 |
| 62199212 | 20000 | 40 | 00:04:34 |
| 62199213 | 50000 | 10 | 00:00:55 |
| 62199214 | 50000 | 20 | 00:00:45 |
| 62199215 | 50000 | 40 | 00:00:44 |
...
Walltime: 00-01:00:00
========================================================================
...
slurmstepd: error: Detected 1 oom_kill event in StepId=62199210.batch.
Some of the step tasks have been OOM Killed.- 1
- Oopsie, Out Of Memory (OOM)
Memory
Check RAM of node in docs, typically 256 GB
- Total memory for job
specify with
--mem, e.g.,--mem< RAM - 8 GB
- Memory per CPU
specify with
--mem-per-cpu, e.g.,--mem-per-cpu\(\times\)--cpus-per-task\(\times\)--ntasks< RAM - 8 GB
Units: K, M, G, T
How much memory do you need?
- Specify memory (
--mem=30) - But how much?
- Don’t underestimate!
- Don’t (massively) overestimate
- Experiment
- Start with “small” problem
- Increase problem size gradually
- Extrapolate to real problem
How much was used?
Submit with range of values for SIZE and POWER, e.g.,
$ sbatch --export=ALL,SIZE=5000,POWER=10 memory_and_walltime.slurm
$ sbatch --export=ALL,SIZE=5000,POWER=20 memory_and_walltime.slurm
...When job, e.g., 62199204, finished, use sacct
Tip
Combine walltime and memory benchmark!
Memory benchmark results
| job ID | \(N\) | \(p\) | \(M\) (GB) |
|---|---|---|---|
| 62204487 | 5000 | 10 | 0.82 |
| 62204488 | 5000 | 20 | 1.01 |
| 62204489 | 5000 | 40 | 0.82 |
| 62204490 | 10000 | 10 | 3.81 |
| 62204491 | 10000 | 20 | 3.82 |
| 62204492 | 10000 | 40 | 3.82 |
| 62204493 | 20000 | 10 | 14.61 |
| 62204494 | 20000 | 20 | 12.03 |
| 62204495 | 20000 | 40 | 12.03 |
- \(M(N, p) \propto N^2\)
- \(M(N, p)\) constant in \(p\)
Summary walltime & memory
Do not underestimate!
Warning
You will waste resource
Benchmark for walltime and memory
Warning
Unless cost of benchmarking is higher than production cost
Efficiency
Implicit parallelism
Some R functions/operations are parallel under the hood
How do you know?
- Check the documentation
- Measure it
Benchmark environment
Experiments done on Sapphire Rapids node
- 2 Intel Xeon Platinum 8468 48-core
- 256 GB RAM
Module R/4.4.0-gfbf-2023a
Parallel or not?
Time the execution of R script
- 1
- Real time, i.e., walltime
- 2
- User time, i.e., CPU time used
82 seconds CPU time in 4.8 seconds walltime… parallelism!
%*% use Basic Linear Algebra Subroutines (BLAS)
Uses multiple threads, each on a core
How to exploit that?
Easy: set --cpus-per-task > 1
| CPUs per task (\(n\)) | walltime (\(T_n\)) |
|---|---|
| 1 | 109.6 |
| 2 | 64.8 |
| 4 | 50.5 |
| 8 | 40.2 |
| 12 | 34.3 |
| 24 | 31.5 |
| 48 | 31.0 |
| 96 | 33.0 |
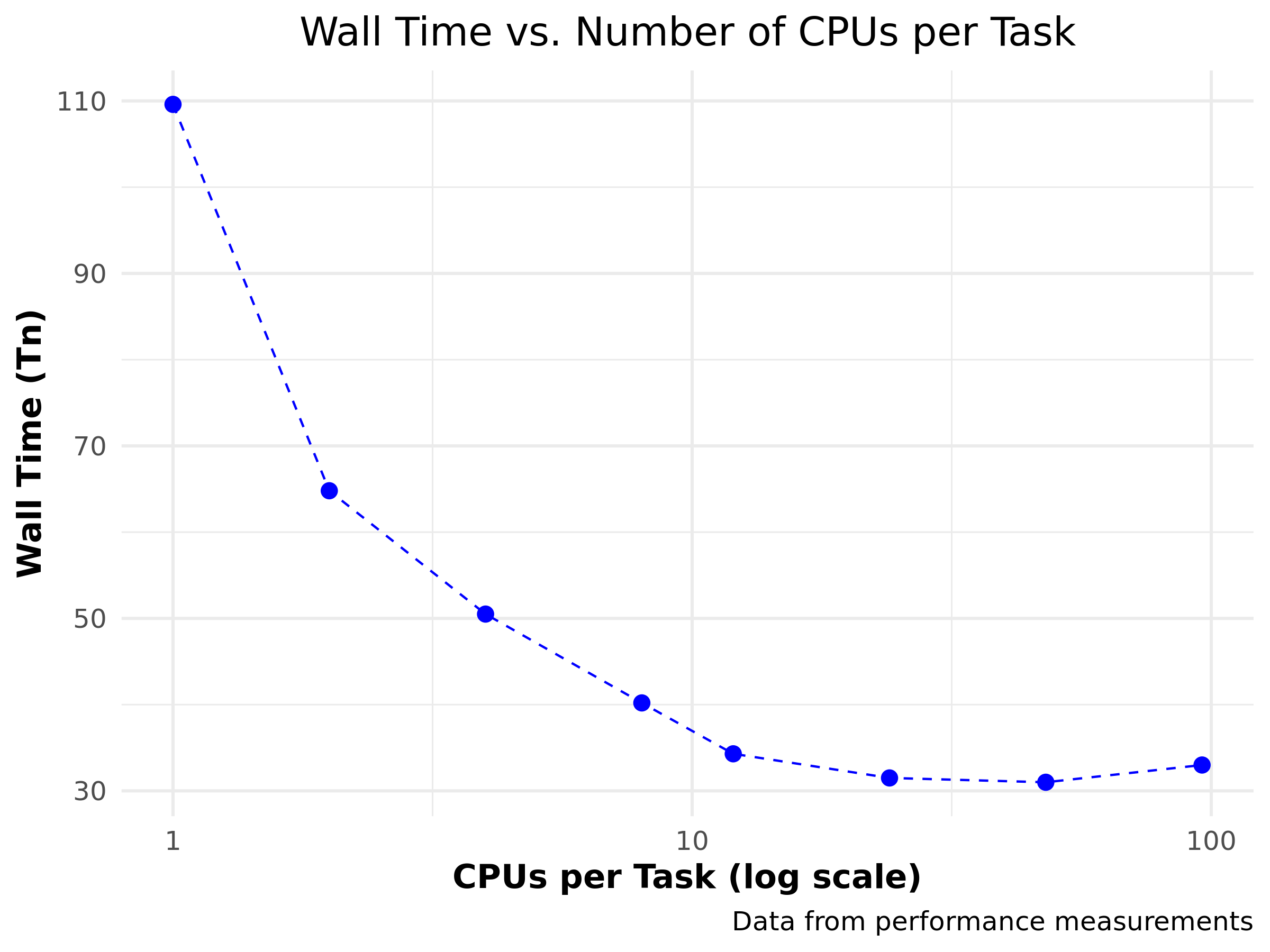
Nice!
Speedup
\(S(n) = \frac{T_1}{T_n}\)
| \(n\) | \(T_n\) | \(S(n)\) |
|---|---|---|
| 1 | 109.6 | 1.00 |
| 2 | 64.8 | 1.69 |
| 4 | 50.5 | 2.17 |
| 8 | 40.2 | 2.73 |
| 12 | 34.3 | 3.20 |
| 24 | 31.5 | 3.48 |
| 48 | 31.0 | 3.54 |
| 96 | 33.0 | 3.32 |
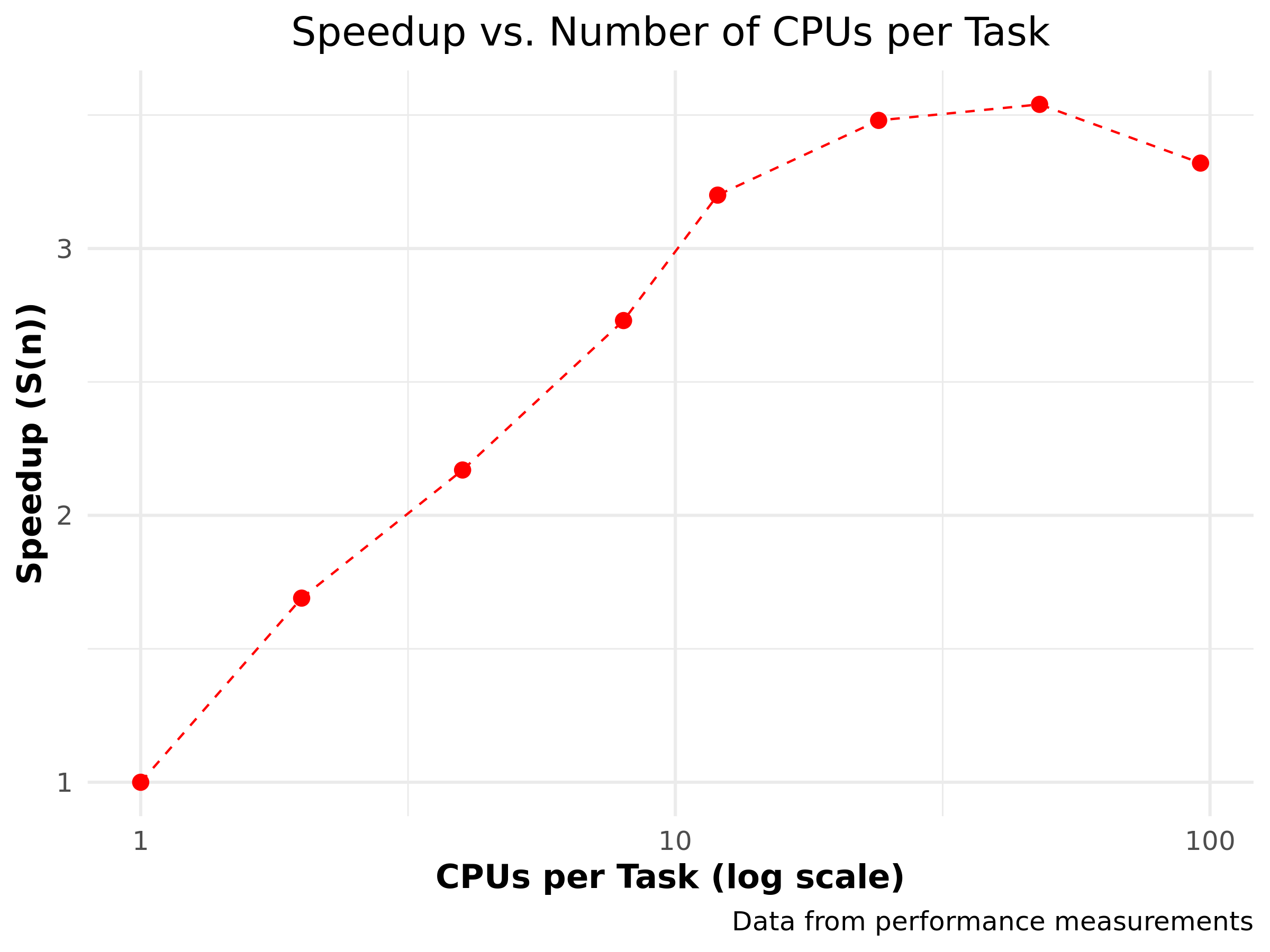
Nice? Maybe…
Parallel efficiency
\(E(n) = \frac{T_1}{n \cdot T_n}\)
| \(n\) | \(T_n\) | \(S(n)\) | \(E(n)\) |
|---|---|---|---|
| 1 | 109.6 | 1.00 | 1.00 |
| 2 | 64.8 | 1.69 | 0.85 |
| 4 | 50.5 | 2.17 | 0.54 |
| 8 | 40.2 | 2.73 | 0.34 |
| 12 | 34.3 | 3.20 | 0.27 |
| 24 | 31.5 | 3.48 | 0.14 |
| 48 | 31.0 | 3.54 | 0.07 |
| 96 | 33.0 | 3.32 | 0.03 |
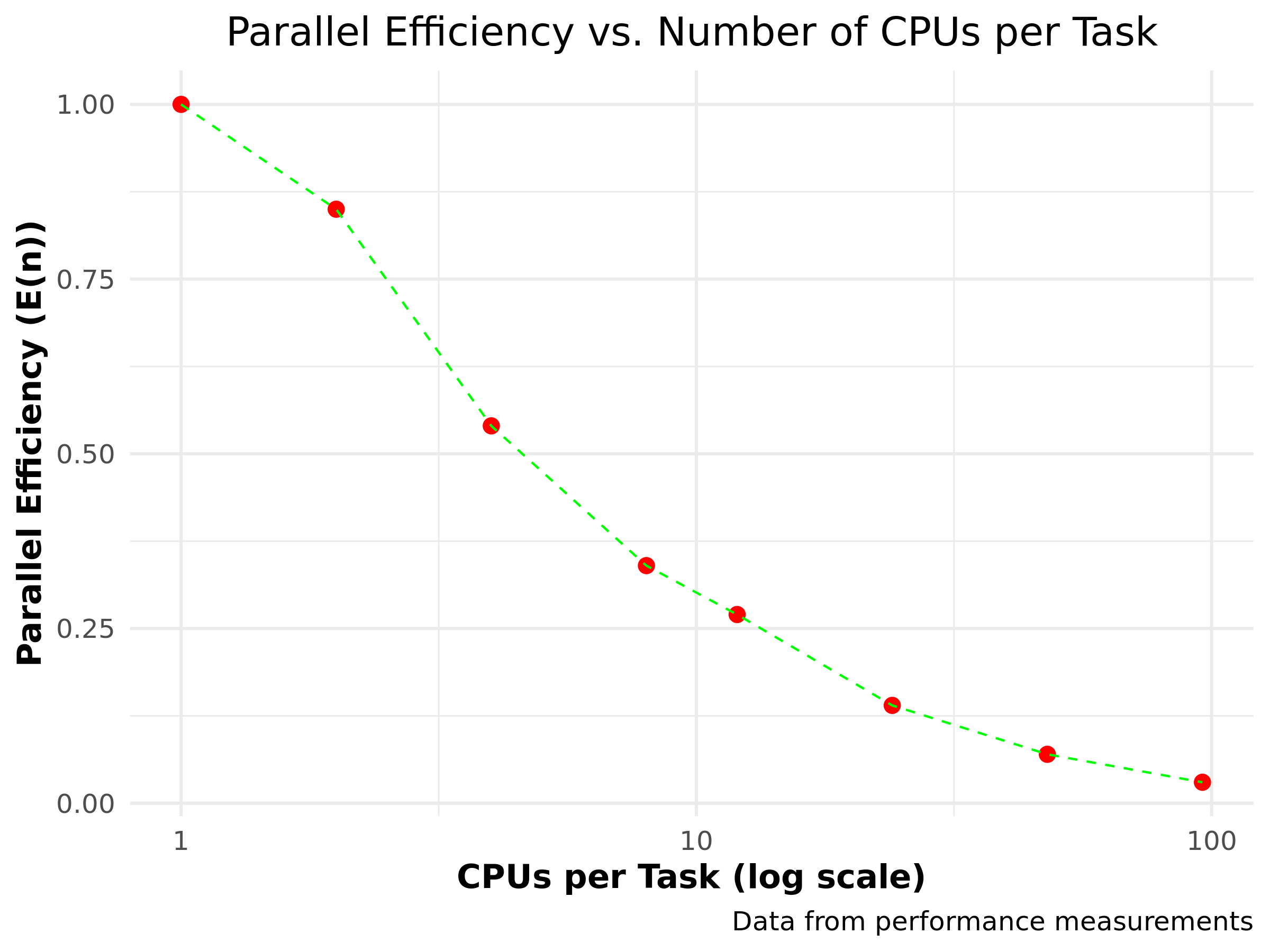
Nice?, not so much… HPC infrastructure is very expensive!
Lots of matrices…
Often same script run for many parameter settings (\(N\))
| \(N\) | \(T_N\) | \(S(N)\) | \(E(N)\) |
|---|---|---|---|
| 1 | 14.0 | 1.00 | 1.00 |
| 2 | 33.6 | 0.42 | 0.21 |
| 4 | 90.1 | 0.16 | 0.04 |
| 8 | 184.4 | 0.08 | 0.01 |
| 12 | 252.5 | 0.06 | 0.005 |
| 18 | 635.2 | 0.02 | 0.001 |
Oops!?! Worse than serial?
All R instances use 96 threads, so for \(N = 18\)… \(18 \times 96 = 1728\) threads \(\gg\) number of cores
Warning
Massive oversubscription
Control OMP_NUM_THREADS!
Set OMP_NUM_THREADS=2
| \(N\) | \(T_N\) | \(S(N)\) | \(E(N)\) |
|---|---|---|---|
| 1 | 42.6 | 1.00 | 1.00 |
| 4 | 43.7 | 3.90 | 0.98 |
| 8 | 49.6 | 6.87 | 0.86 |
| 12 | 65.3 | 7.82 | 0.65 |
| 24 | 74.5 | 13.7 | 0.57 |
| 36 | 85.2 | 18.0 | 0.50 |
| 48 | 94.3 | 21.6 | 0.45 |
\(N = 8\) seems optimal, but what the heck, you’re ruining the node anyway
Better go all the way to \(N = 48\)
Tip
OMP_NUM_THREADS \(\le\) number of cores
The good news is…
Applications can be
- Memory bound 😒
- CPU bound 😊
- I/O bound 😗
Warning
Benchmark you code!
Why performance degradation?
It’s the memory architecture, stupid!
- 48 core Intel Sapphire Rapids CPU
- 256 GB RAM, 8 memory channels at 4800 MHz
- L3 cache per socket: 105 MB (shared, 2.2 MB per core)
- L2 cache per CPU (core): 2 MB
- L1 cache per CPU (core): 48 kB data
- 36 core Intel Icelake
- 256 GB RAM, 8 memory channels at 3200 MHz
- L3 cache per socket: 54 MB (shared, 1.5 MB per core)
- L2 cache per CPU (core): 1.25 MB
- L1 cache per CPU (core): 48 kB data
How to time?
In job script:
timeIn job script:
hyperfine- Runs command multiple times
- Reports mean, median, min, max, etc.
- Can compare multiple commands
For completed job
And another thing…
Reading/writing many small files: bad idea!
Warning
File systems for HPC optimized for large read/write operations, not for many metadata operations!
Use appropriate file types, e.g.,
- HDF5
- Parquet
Improving performance
Where to start?
- Gut feeling?
- Best guess?
- Easy to optimize?
Tip
To measure is to know: use a profiler
Profiling
- R has
RprofandsummaryRprof- Easy to use
- wrap around R script using
source
Profile
Code optimization: algorithm first!
%*%pretty good, not much chance- However, \(A^p = A \cdot \cdots \cdot A\)?
- \(p - 1\) multiplications
Think in terms of the algorithm
\[ A^{13} = A \cdot A^4 \cdot A^8 \]
Binary exponentiation: 5 versus 12 muliplications
Wrap up
Conclusions
- Running R on HPC is easy
- Running R on HPC efficiently… requires care
How to proceed?
If necessary, take trainings
- HPC intro
- Linux intro
- Contact support
Tip
If you are a programmer, consider parallelization